-
GTD-211
GLUTAMATE DEHYDROGENASE (NAD-dependent) from Microorganism

PREPARATION and SPECIFICATION
| Appearance | White amorphous powder, lyophilized | ||
|---|---|---|---|
| Activity | GradeⅡ 100 U/mg-solid or more | ||
| Contaminants | NADH oxidase | ≤1.0×10-2% | |
PROPERTIES
| Stability | Stable at −20℃ for at least one year(Fig.1) |
|---|---|
| Molecular weight | approx. 260,000 |
| Isoelectric point | 5.6 |
| Michaelis constants | 9.21×10-3M (NH3), 4.80×10-3M(α-Ketoglutarate) 7.8×10-5M (L-Glutamate), 1.29×10-4M(NADH), 5.89×10-4M(NAD+) |
| Structure | 6 subunits per enzyme molecule |
| Inhibitors | Heavy metals, PCMB, IAA |
| Optimum pH | 7.5−8.0 (α-KG→L-Glu) 9.0 (L-Glu→α-KG)(Fig.2) |
| Optimum temperature | 55℃ (α-KG→L-Glu) 50℃ (L-Glu→α-KG)(Fig.3) |
| pH Stability | pH 5.0−10.0 (25℃, 20hr)(Fig.4) |
| Thermal stability | below 50℃ (pH 8.3, 10min)(Fig.5) |
| Substrate specificity | (Table 1) |
| Effect of various chemicals | (Table 2) |
APPLICATIONS
This enzyme is useful for enzymatic determination of NH3, α-ketoglutaric acid and L-glutamic acid, and for assay of leucine aminopeptidase and urease. This enzyme is also used for enzymatic determination of urea when coupled with urease (URH-201) in clinical analysis.
ASSAY
Principle

The disappearance of NADH is measured at 340nm by spectrophotometry.
Unit definition
One unit causes the oxidation of one micromole of NADH per minute under the conditions described below.
Method
Reagents
| A. Buffer solution | 0.1M Tris-HCl buffer, pH 8.3 | |
|---|---|---|
| B. NH4Cl solution | 3.3M | |
| C. α-Ketoglutarate solution | 0.225M (adjust the pH to 7.0−9.0 with NaOH)(Should be phepared fresh) | |
| D. NADH solution | 7.5mM (Should be prepared fresh) | |
| E. Enzyme diluent | 0.1M Tris-HCl buffer, pH 8.3 | |
Procedure
1. Prepare the following reaction mixture in a cuvette (d=1.0cm) and equilibrate at 30℃ for about 5 minutes.
| 2.5ml | Buffer solution | (A) |
| 0.2ml | NH4Cl solution | (B) |
| 0.1ml | α-Ketoglutarate solution | (C) |
| 0.1ml | NADH solution | (D) |
| Concentration in assay mixture | |
|---|---|
| Tris-HCl buffer | 86 mM |
| α-Ketoglutarate | 7.6 mM |
| NH4Cl | 0.22 M |
| NADH | 0.25mM |
2. Add 0.05ml of the enzyme solution* and mix by gentle inversion.
3. Record the decrease in optical density at 340nm against water for 2 to 3 minutes in a spectrophotometer thermostated at 30℃, and calculate the ΔOD per minute from the initial linear portion of the curve (ΔOD test).
At the same time, measure the blank rate (ΔOD blank) by using the same method as the test except that the enzyme diluent (E) is added instead of the enzyme solution.
*Dissolve the enzyme preparation to 0.1−0.8U/ml with ice-cold diluent (E), immediately before assay.
Calculation
Activity can be calculated by using the following formula :
Volume activity (U/ml) =
-
ΔOD/min (ΔOD test−ΔOD blank)×Vt×df
6.22×1.0×Vs
= ΔOD/min×9.486×df
Weight activity (U/mg) = (U/ml)×1/C
| Vt | : Total volume (2.95ml) |
| Vs | : Sample volume (0.05ml) |
| 6.22 | : Millimolar extinction coefficient of NADH at 340nm (cm2/micromole) |
| 1.0 | : Light path length (cm) |
| df | : Dilution factor |
| C | : Enzyme concentration in dissolution (c mg/ml) |
Table 1. Substrate Specificity of Glutamate dehydrogenase
-
Substrate (2mM) Relative activity(%) L-Glutamate 100 L-Norvaline 0.35 L-α-Aminobutyrate 0.16 L-Norleucine 0 D,L-Homocysteine 0.06 L-Isoleucine 0.09 -
Substrate (2mM) Relative activity(%) L-Glutamine 0.05 L-Aspartate 0.07 L-Asparagine 0.11 L-Valine 0.09 L-Leucine 0.03 L-Alanine 0.07 L-Methionine 0.06
Glutamate dehydrogenase : 0.3U/ml of 0.1M Tris-HCl buffer, pH 9.0 NAD+:12mM
Table 2. Effect of Various Chemicals on Glutamate dehydrogenase
[The enzyme dissolved in 0.1M Tris-HCl buffer, pH 8.3 was incubated with each chemical at 25℃ for 1hr.]
-
Chemical Concn.(mM) Residual
activity(%)None - 100 Metal salt 2.0 MgCl2 97 CaCl2 99 Ba(OAc)2 101 FeCl3 1.8 CoCl2 97 MnCl2 78 ZnSO4 6.9 Cd(OAc)2 58 NiCl2 100 CuSO4 0.3 Pb(OAc)2 0.01 AgNO3 1.6 HgCl2 0 PCMB 2.0 0.6 MIA 2.0 98 -
Chemical Concn.(mM) Residual
activity(%)NaF 2.0 100 NaN3 20 102 EDTA 5.0 102 o-Phenanthroline 2.0 101 α,α′-Dipyridyl 2.0 102 Borate 102 IAA 2.0 0.2 NEM 2.0 96 Hydroxylamine 2.0 100 Triton X-100 0.10% 102 Brij 35 0.10% 103 Tween 20 0.10% 101 Span 20 0.10% 107 Na-cholate 0.10% 103 SDS 0.05% 0.1 DAC 0.05% 0.2
Ac, CH3CO; PCMB, p-Chloromercuribenzoate; MIA, Monoiodoacetate; NEM, N-Ethylmaleimide; IAA, Iodoacetamide; EDTA, Ethylenediaminetetraacetate; SDS, Sodium dodecyl sulfate; DAC, Dimethylbenzylalkylammonium chloride
-
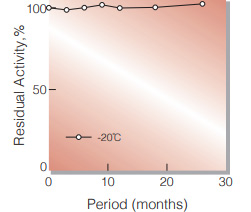
Fig.1. Stability (Powder form)
(kept under dry conditions)
-
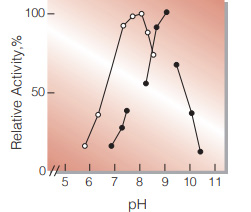
Fig.2. pH-Activity
○̶○,α-KG →L-Glu;●̶●L-Glu →α-KG in 0.1M buffer solution:pH5.7-7.6 K-phosphate,pH7.8-9.0,Tris-HCI; pH9.4-10.3, glycine-NaOH
-
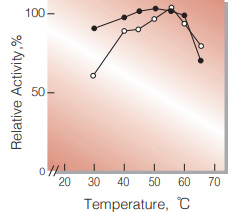
Fig.3. Temperature activity
○̶○,α-KG →L-Glu;0.1M Tris-HCI buffer pH8.3;●̶●,L-Glu →α-KG:0.1M Tris-HCI buffer,pH9.0
-
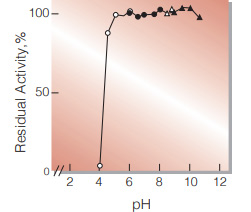
Fig.4. pH-Stability
25℃, 20hr-treatment with 0.1M buffer solution: ○̶○,acetate;●̶●,K-phosphate,△̶△Tris-HCI;▲̶▲glycine-NaOH
-
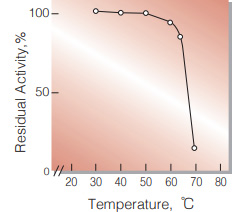
Fig.5. Thermal stability
10min-treatment with 0.1M Tris-HCI buffer, pH8.3
活性測定法(Japanese)
1. 原理

NADHの消失量を340nmの吸光度の変化で測定する。
2.定義
下記条件下で1分間に1マイクロモルのNADHが酸化される酵素量を1単位(U)とする。
3.試薬
- 0.1M Tris-HCl緩衝液, pH8.3
- 3.3M NH4Cl水溶液
- 0.225M α-ケトグルタル酸水溶液(NaOHでpHを7.0〜9.0に調整)(用時調製)
- 7.5mM NADH水溶液(用時調製)
酵素溶液:分析直前に酵素標品を予め氷冷した0.1M Tris-HCl緩衝液,pH8.3で0.1~0.8U/㎖に希釈する。
4.手順
1.下記反応混液をキュベット(d=1.0cm)に調製し,30℃で約5分間予備加温する。
| 2.5ml | Tris-HCl緩衝液 | (A) |
| 0.2ml | NH4Cl水溶液 | (B) |
| 0.1ml | α-ケトグルタル酸水溶液 | (C) |
| 0.1ml | NADH水溶液 | (D) |
2.酵素溶液を0.05㎖を添加し,ゆるやかに混和後,水を対照に30℃に制御された分光光度計で340nmの吸光度変化を2~3分間記録し,その初期直線部分から1分間当りの吸光度変化を求める(ΔODtest)。
3.盲検は反応混液①に酵素溶液の代りに酵素希釈液(0.1M Tris-HCl緩衝液,pH8.3)を加え,上記同様に操作を行って,1分間当りの吸光度変化を求める(ΔODblank)。
5.計算式
U/ml =
-
ΔOD/min (ΔOD test−ΔOD blank)×2.95(ml)×希釈倍率
6.22×1.0×0.05(ml)
| = ΔOD/min×9.486×希釈倍率 | |
| U/mg | = U/ml×1/C |
| 6.22 | : NADHのミリモル分子吸光係数(cm2/micromole) |
| 1.0 | : 光路長(cm) |
| C | : 溶解時の酵素濃度(c mg/ml) |
CONTACT
お問い合わせ-
各種製品に関するご質問・ご相談はこちらよりお問い合わせください。
